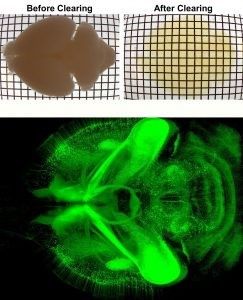
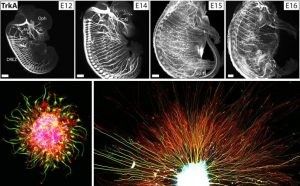
In the Wu Lab, we work to study and document the structures of neural systems at the whole organ and organism levels. Our study is guided by the questions:
“How do diverse neural circuits precisely form during development?”
&
“How are neural circuits disrupted by genetic mutations or disease?”
It is commonly known that complex neural circuits form within the brain to control basic biological processes and behavior. Similarly, neurons’ axons innervate varied organs in the peripheral nervous system, carrying signals that regulate the organs’ physiological functions. Understanding the architecture of these neural structures is important to understanding their function. Yet capturing a complete architecture poses unique challenges. When a single neuron’s numerous, sub-micron thin projections can extend across the entire brain and beyond in multiple directions, how can that neuron and its associated circuitry be investigated in detail in its entirety?
To tackle this challenge, we develop and apply novel three-dimensional imaging methods in our lab (the iDISCO family of tissue clearing) which we combine with genetic approaches. This platform allows us to image intact organs and embryos to examine their internal neural structures in true three dimensions. With this platform, we study how neural circuits assemble in the developing mouse embryo. This model helps us answer questions about how healthy circuitry is established and how neural wiring can go wrong. We also study adult animals to characterize the fully established circuitry and to investigate how ongoing changes in circuitry structure may play a role in aging and disease processes. Understanding the structural changes that occur will hopefully give us insights into how we may restore function in human individuals suffering from genetic mutations or disease.
We are a vibrant, inclusive, and synergistic team with multidisciplinary expertise (genetics and modern histology, cellular and molecular biology, advanced volumetric imaging, and big-data analysis using machine learning), to facilitate our highly collaborative research spanning basic systematic profiling of the brain to pathological discovery with animal models and human patient samples. Our motivation is to contribute to the development of both tools/resources and biological insights, as well as to engage in community effort for the education and training of scientists to lead future brain research.